introduction
The RNA pull-down technique, as a key method for exploring the interaction between RNA and proteins, is receiving widespread attention. RNA pull-down is an in vitro RNA pull-down technique, which allows the identification of protein partners that interact specifically with the RNA molecules of interest. Its principle is based on using biotin-labeled RNA probes as “baits” to capture the proteins that bind specifically to them in the cell lysate.
advantages
✔ It can specifically obtain the interacting proteins of an RNA (including long non-coding RNA (lncRNA), circular RNA (circRNA), microRNA (miRNA), specific transcripts of mRNA, etc.), and analyze the molecular mechanisms by which RNA exerts its functions and roles.
✔ There are two supporting experimental schemes available: the full-length method and the probe method. These can achieve the capture of all types of RNA molecules, eliminating limitations such as length and category restrictions, so as to accurately obtain the proteins captured by the target gene and exclude non-specific interference.
✔ The experimental operation process is simple, with a high success rate and a controllable cycle.
principle
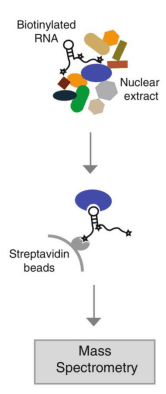
This technique involves using in vitro transcription technology to prepare biotin-labeled RNA probes, and then mixing them with cytoplasmic protein extracts to form RNA-protein complexes. These complexes can bind to streptavidin-modified magnetic beads, enabling separation from other components in the mixture. After the complexes are eluted, Western blot (WB) experiments can be used to confirm whether specific RNA-binding proteins have interacted with the RNA, or mass spectrometry (MS) technology can be utilized to screen for unknown proteins that bind to the RNA.
Currently, there are two methods for our RNA pull down:
1) Full-length method:
The full-length sense strand-biotin-labeled probe is used to “fish” for the bound proteins, while the full-length antisense strand-biotin-labeled probe is captured as the control group. The full-length method uses the entire gene as a probe to “fish”, which is equivalent to the capture of the binding between exogenous genes and endogenous proteins.
2) Probe method:
When the gene is relatively long, for example, when its length exceeds 3 kb, or when targeting a specific transcript and avoiding other transcripts, the probe method will be used. The short fragment-antisense strand-biotin-labeled probe binds to the target gene of the sense strand in vivo to “fish” for the proteins bound to the target gene in vivo. The short fragment-antisense strand-unlabeled probe (without biotin labeling) is used as the control group, which is equivalent to the capture of the proteins bound to the endogenous target gene.
Demo Data

The results of the SDS-PAGE gel of proteins bound to lincRNA-p21 (right lane) or antisense RNA (left lane) are presented. hnRNP-K was identified as a band specific to lincRNA-p21. Western blot analysis of the specific binding between hnRNP-K and lincRNA-p21 also showed a non-specific protein (NONO) as a control.
Research Cases
Case 1
Title: A novel antiviral long non-coding RNA (lncRNA), EDAL, shields a T309 O-GlcNAcylation site to promote lysosomal degradation of EZH2.
Journal: Genome Biology
Impact Factor: 10.1

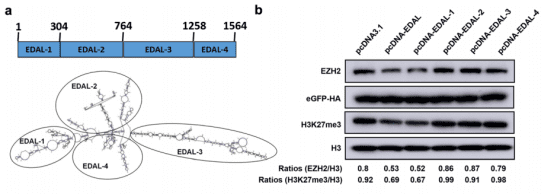
Professor Zhao Ling from Huazhong Agricultural University and his team have revealed the inhibitory function of a long non-coding RNA (lncRNA) called EDAL in the nervous system against neurotropic viruses including the rabies virus, opening up new hopes for the treatment after rabies onset. The study pointed out that neurotropic viruses can stimulate the expression of lncRNA EDAL in nerve cells. EDAL can specifically bind to EZH2 and promote the degradation of EZH2 through the lysosomal pathway by regulating the O-GlcNAcylation modification at the T309 site of EZH2. This process ultimately leads to the expression of the antiviral polypeptide PCP4L1, thereby inhibiting the proliferation of multiple neurotropic viruses. This study provides a new perspective for in-depth exploration of the mechanism of interaction between neurotropic viruses and the host.
To deeply investigate which sequence segment in EDAL is crucial for its antiviral function, on the basis of analyzing the structure of EDAL, the researchers divided it into four parts and examined their impacts on the degradation of EZH2 one by one. The results of RNA pull-down showed that only EDAL-1 could significantly reduce the levels of EZH2 and H3K27me3 as well as the proliferation of the rabies virus. By removing different sequences in EDAL-1, the researchers determined that the sequence with a length of 56 bases located between positions 98 and 153 might be the core of its antiviral activity. When this sequence was integrated into the structures of other long non-coding RNAs (lncRNAs), antiviral effects were also observed.
Case 2
Title: Long noncoding RNA DLGAP1-AS2 Promotes Tumorigenesis and Metastasis by Regulating the Trim21/ELOA/LHPP Axis in Colorectal Cancer
Journal: Molecular Cancer
Impact Factor: 27.7

The research team from the Affiliated Hospital of Jiangnan University has revealed the great potential of DLGAP1-AS2 as a prognostic biomarker. This discovery has opened up a new perspective for the research on the molecular mechanisms of colorectal cancer and provided new targets for treatment strategies.
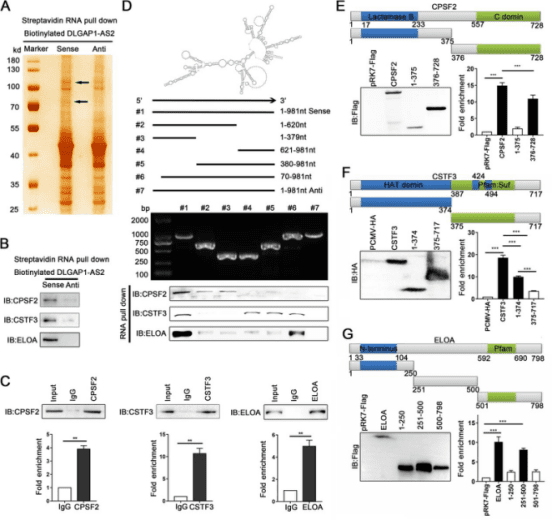
Firstly, the study conducted transcriptome analysis on colorectal cancer and its adjacent tissues by using next-generation sequencing technology, and identified a series of long non-coding RNAs with significant differences in expression in colorectal cancer. Through the analysis of public databases and the verification of clinical samples, the researchers selected DLGAP1-AS2, which was significantly upregulated in tumors, as the focus of the study. Clinical data showed that the expression level of DLGAP1-AS2 in colorectal cancer was increased, which might be driven by the increase in DNA copy number, and its high-level expression was closely associated with the poor prognosis of patients. Further studies identified a new dominant transcript (MK336171) of DLGAP1-AS2 in colorectal cancer, and found through in vitro and in vivo experiments that DLGAP1-AS2 could promote the growth and metastasis of colorectal cancer.
Mechanistically, through techniques such as RNA pull-down and RNA immunoprecipitation (RIP), the researchers identified the binding proteins CPSF2, CSTF3 and ELOA that interact with DLGAP1-AS2. It was revealed that DLGAP1-AS2 promoted the interaction between the E3 ubiquitin ligase Trim21 and the transcription elongation factor A (ELOA), increased the ubiquitin-mediated degradation of ELOA, thereby inhibiting its transcriptional activation of the downstream target gene LHPP, leading to the activation of the AKT signaling pathway and further promoting tumorigenesis and development. In addition, the study also confirmed that CPSF2 and CSTF3 could bind to DLGAP1-AS2 and work together to enhance its stability.
Case 3
Title: CircEAF2 Counteracts Epstein-Barr Virus-Positive Diffuse Large B-Cell Lymphoma Progression via the miR-BART19-3p/APC/β-catenin Axis
Journal: Molecular Cancer
Impact Factor: 27.7

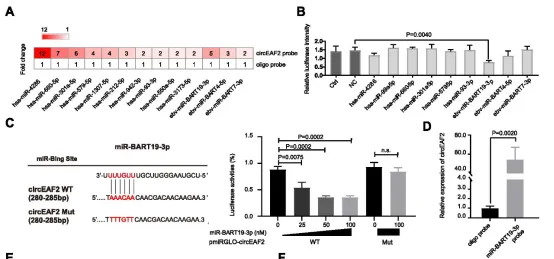
The formation of lymphoma is closely related to Epstein-Barr virus (EBV), especially significantly associated with the poor prognosis of diffuse large B-cell lymphoma (DLBCL). Circular RNA (circRNA) plays a crucial role in the development process of lymphoma. Nevertheless, we still know very little about the potential mechanism of action of circRNA in the progression of EBV-related DLBCL. In this study, high-throughput sequencing was performed on tumor samples from 12 DLBCL patients to screen for circRNAs. Subsequently, the expression level of circEAF2 and its relationship with clinical features and prognosis were thoroughly investigated in tumor samples from 100 DLBCL patients by using real-time quantitative PCR technology. The research results revealed a newly discovered circRNA, circEAF2, which had a decreased expression in EBV-positive DLBCL and was negatively correlated with EBV infection and the progression of DLBCL. In EBV-infected B lymphoma cells, the overexpression of circEAF2 could promote apoptosis and enhance the sensitivity of cells to the chemotherapeutic drug epirubicin.
Through RNA pull-down experiments, the study found that circEAF2 specifically targeted the EBV-encoded miR-BART19-3p, upregulated the APC protein, and inhibited the expression of β-catenin, thereby leading to the inactivation of the Wnt signaling pathway and further inhibiting the proliferation of EBV-positive DLBCL cells. In the EBV-positive B lymphoma mouse model, the transplanted tumors with circEAF2 overexpression showed a reduction in Ki-67 positive cells, an increase in apoptosis, and a slowdown in tumor growth. In summary, this study found that circEAF2 inhibits the progression of EBV-positive DLBCL through the miR-BART19-3p/APC/β-catenin axis. Therefore, circEAF2 is regarded as a potential prognostic biomarker. Therapeutic strategies targeting EBV-encoded miRNAs may become a promising direction for the treatment of EBV-related lymphoid malignancies.
