introduction
During the transcription process in eukaryotes, a series of modifications will be carried out on the transcribed RNA, such as adding a cap at the 5′ end and a polyadenylate (Poly(A)) tail at the 3′ end. This structure is of great importance for the stability of mRNA and protein translation.
In 1971, Edmonds discovered that mRNA has a poly(A) tail. It is a sequence of bases added in a non-template-dependent manner by multiple regulatory factors/enzymes at the 3′ end of RNA after transcription, and most of these bases are adenine (A). The poly(A) tail is an essential structural component of the vast majority of eukaryotic mRNA and long non-coding RNA, and it is crucial for RNA transport, stability, translation, and so on.
Poly(A)-seq, namely Poly(A) tail length sequencing, is a technique that can measure the length of poly(A) tails at the whole-genome level. The downstream of the 3′ untranslated region of most eukaryotic mRNA contains a non-templated poly(A) tail, which helps to stabilize the mRNA and transport it to the cytoplasm. The length of the poly(A) tail can affect the translation efficiency of mRNA. Therefore, Poly(A)-seq can be used as a potential tool to analyze the dynamic regulatory processes of mRNA degradation and translation, and it can also discover some unknown characteristics of RNA cleavage and tailing.
advantages
✔ Direct and convenient
✔ Easy to achieve
✔ Low cost
✔ High throughput
✔ High reliability
✔ Rich in information
Applications
1. Affect the translation efficiency of mRNA and protein localization.
2. Participate in the metabolism, transport, decay, and nuclear export of mRNA.
3. Plant defense mechanisms.
4. Targets of anti-tumor drugs.
5. The combined use of PolyA-seq and Ribo-seq can explore the regulatory relationship between the changes in the mRNA polyA tail and the translation efficiency.
6. The combined use of PolyA-seq and RNA-seq can explore the relationship between the changes in the polyA tail and the stability of mRNA.
workflow
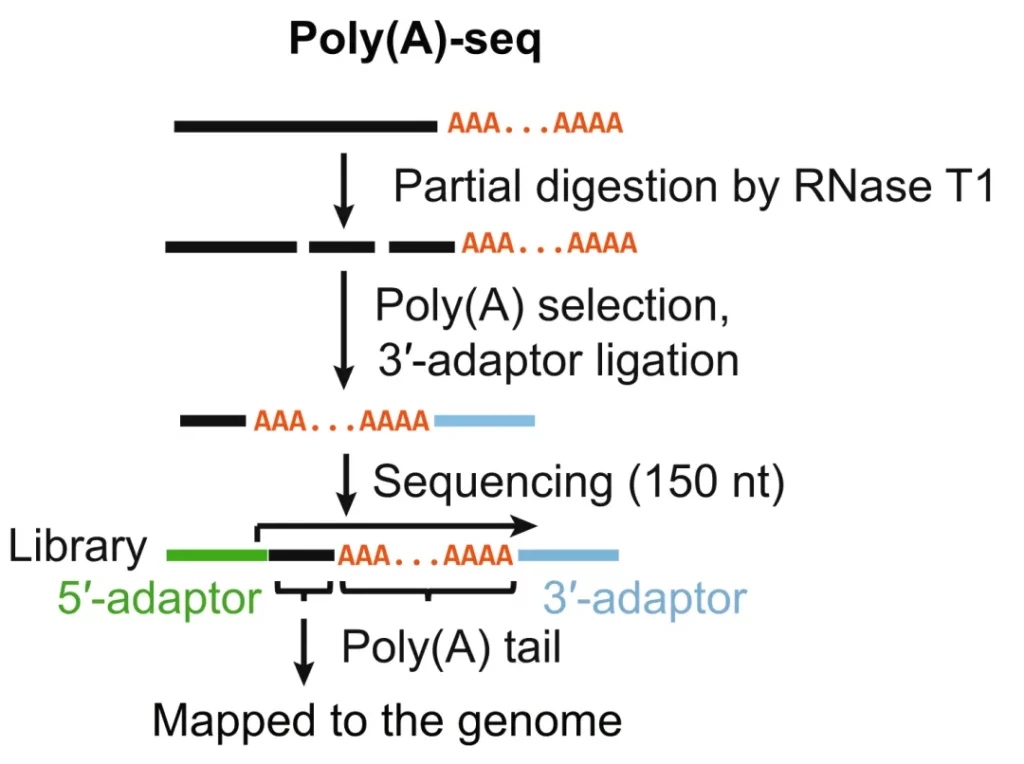
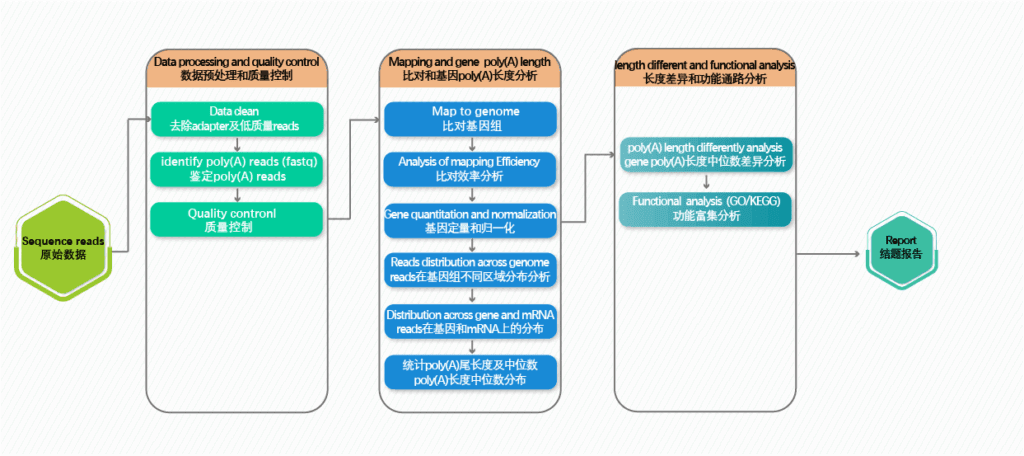
Research Cases
This technique was applied in the research on the poly(A) tailing regulation project of Arabidopsis thaliana, revealing a brand-new post-transcriptional regulation mechanism mediated by the poly(A) tail of Arabidopsis thaliana—sporadically distributed guanylic acid (G) in the poly(A) tail can reduce the translation efficiency of mRNA by inhibiting the interaction with poly(A)-binding protein (PAB). The research findings were published in Genome Biology (Zhao et al., 2019).

In this project, by sequencing the full-length poly(A) tails and developing downstream bioinformatics algorithms to extract high-quality sequencing information, it was found that non-A nucleotides also exist in the poly(A) tails of Arabidopsis thaliana, and the content of G is also the highest among them. Subsequently, the researchers took the core members of the Arabidopsis thaliana poly(A)-binding protein family, namely AtPAB2, AtPAB4 and AtPAB8, as the research objects and constructed a series of important mutants.

Furthermore, by combining high-throughput experimental techniques such as CLIP-seq, ribo-seq, and mRNA stability detection, a systematic study on the molecular biological functions of the poly(A) tail and its binding protein PAB was conducted at the whole-genome level, and the following conclusions were drawn:
1. AtPAB widely binds to the poly(A) tails of mRNA in plants, but there are significant differences in the binding efficiency to different mRNAs.
2. 10% of the poly(A) tails in Arabidopsis thaliana contain at least one guanosine (G), and its content ranges from 0.8% to 28% in different mRNAs.
3. The differential binding of AtPAB to different mRNAs can be partly explained by the differences in the G content in their poly(A) tails—the higher the “purity” of A, the stronger its binding to AtPAB.
4. The binding of AtPAB to mRNA can improve the translation efficiency of mRNA.
5. Correspondingly, in the atpab mutants, the translation efficiency of mRNA with a “pure poly(A) tail” containing no G is reduced more significantly.
6. The homozygous triple mutants of atpab2, atpab4, and atpab8 are lethal, and the double mutants of atpabs exhibit multiple abnormal developmental phenotypes, suggesting that the translation regulation mechanism mediated by the poly(A) tail and its binding protein plays a crucial regulatory role in the normal growth and development of plants.
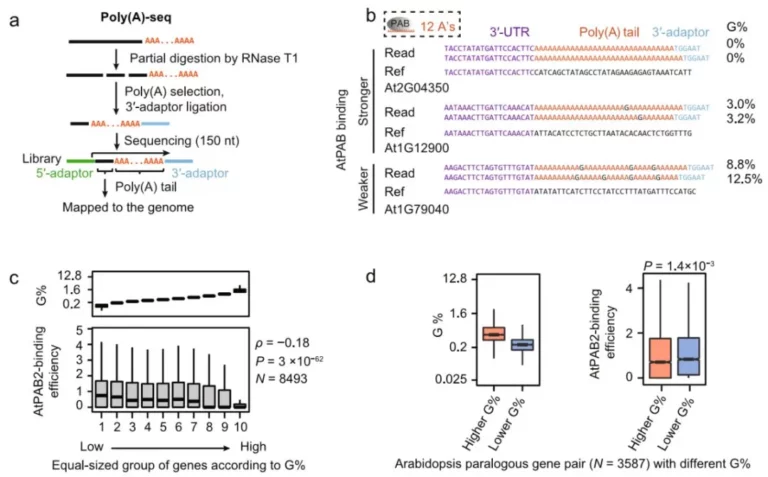
This study fully demonstrated the powerful advantages of the combination of next-generation sequencing technology and bioinformatics algorithms in analyzing the regulatory processes of biological macromolecules. Its results clarified that the G content in the poly(A) tails of plants can affect the protein translation efficiency by inhibiting PAB binding, which is an expansion and innovation of the central dogma of modern molecular biology. This study has important reference value for exploring the post-transcriptional regulatory mechanisms of mRNA in other species.
