RXBio Translates Sequence to Science and Industry
Tel: 027-87050299Email: sales@rxbio.cc
- Home
- Single-cell Sequencing
- Spatial Transcriptomics
- Third-generation Sequencing
- Omics Technologies
- Bioinformatics
- Experimental Platform
- About US
中文
RXBio Translates Sequence to Science and Industry
AbSeq ( also known as CITE-seq) , a single-cell proteomics technology, is an innovative single-cell multi-omics technology developed by BD Biosciences. It utilizes oligonucleotide-labeled antibodies to achieve digital detection of protein expression and integrates with the BD Rhapsody™ system to enable simultaneous quantitative analysis of mRNA and proteins at the single-cell level. AbSeq provides powerful support for in-depth analysis of cell heterogeneity, phenotypic and functional states, and is especially applicable to research on immune cells.
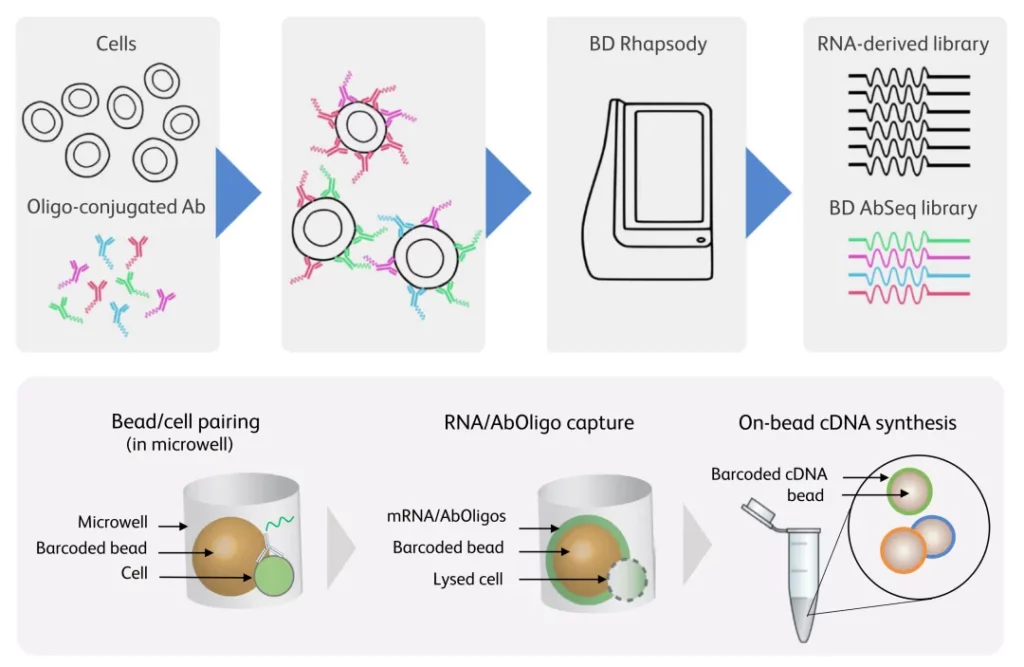
Use oligonucleotide-conjugated antibodies (AbSeq antibodies) to specifically bind to single cells, and obtain protein expression information at the single-cell level through next-generation sequencing technology.
01 Is cell heterogeneity hard to understand?
Use oligonucleotide-conjugated antibodies (AbSeq antibodies) to specifically bind to single cells, and obtain protein expression information at the single-cell level through next-generation sequencing technology.
02 Are rare immune cells difficult to find?
The high-sensitivity detection ability of AbSeq helps with the effective capture and analysis of rare cell types such as low-frequency immune cells.
03 Is it difficult to detect low-abundance proteins?
AbSeq significantly improves the detection ability of low-expressing proteins and overcomes the limitations of traditional methods.
04 Looking for therapeutic targets?
The expression of mRNA and proteins is not always consistent. AbSeq provides comprehensive multi-dimensional information at the single-cell level and accelerates the identification of therapeutic targets and disease markers.
05 Is it hard to track cell dynamics?
AbSeq can simultaneously analyze protein expression and gene transcription at the single-cell level, revealing the dynamic changes of cell responses.
06 Studying cell interactions?
AbSeq, combined with information on cell surface proteins, helps to clarify the communication networks among cells and their roles in biological processes.
AbSeq on the BD Rhapsody™ system is superior to antibodies of other brands in terms of resolution. Select antibodies with the same clone from BD and competing products, and use BD AbSeq or competing products at the same concentration to incubate cells from the same source and in the same quantity. The AbSeq on the BD Rhapsody™ system is significantly better than competing products in terms of resolution.
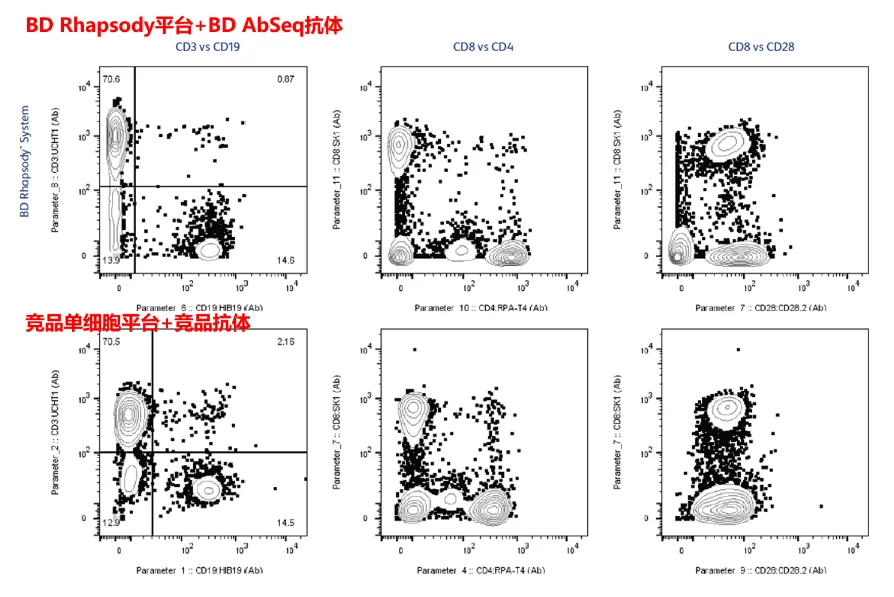
The BD Rhapsody™ system demonstrates a higher resolution and clearly shows cell populations with high, medium, and negative expressions. (For example, as the expression of CD28 is known to be relatively low, the single-cell system of competing products is unable to distinguish different CD28 negative/positive populations.)
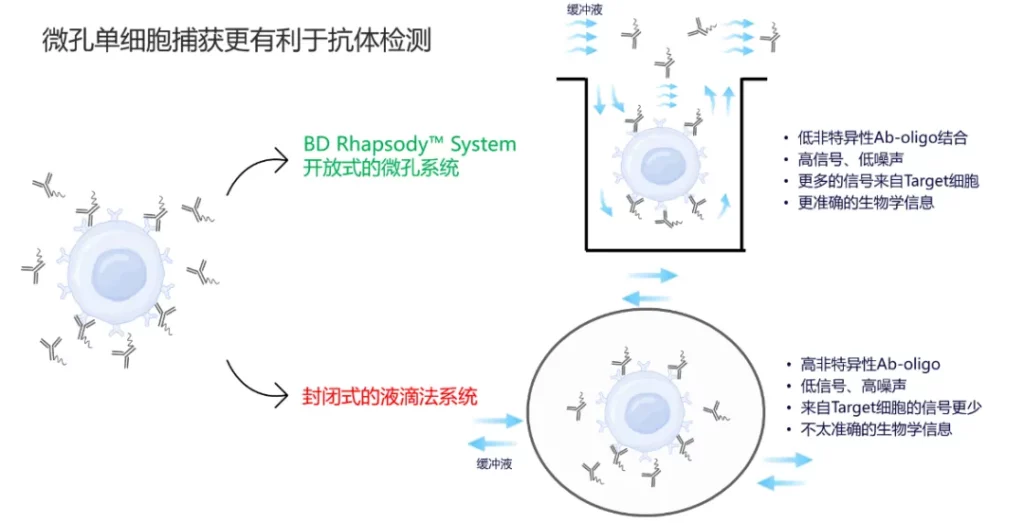
In BD’s single-cell microwell separation and capture method, Ab-oligo antibodies that have not bound to the target cells are more likely to be completely washed away during multiple wash steps. This ensures that all the antibodies detected subsequently are those that have bound to the target cells, resulting in a higher signal-to-noise ratio and the revelation of more accurate biological information.
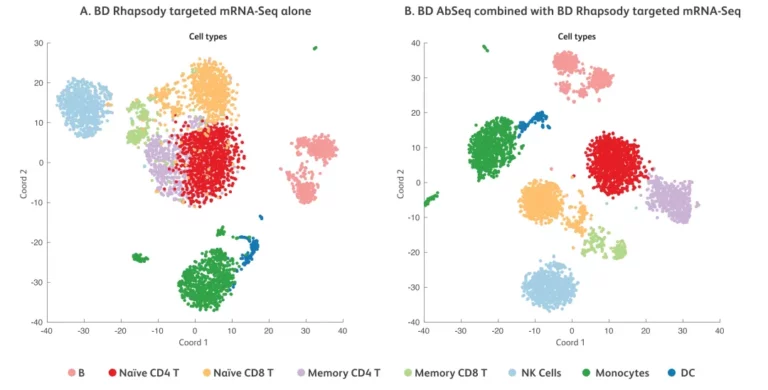
When relying solely on mRNA expression data (Figure A), the clustering of cell types is relatively ambiguous. After adding protein expression data (Figure B), the clustering of cell types becomes clearer and the boundaries between different cell types are more distinct.
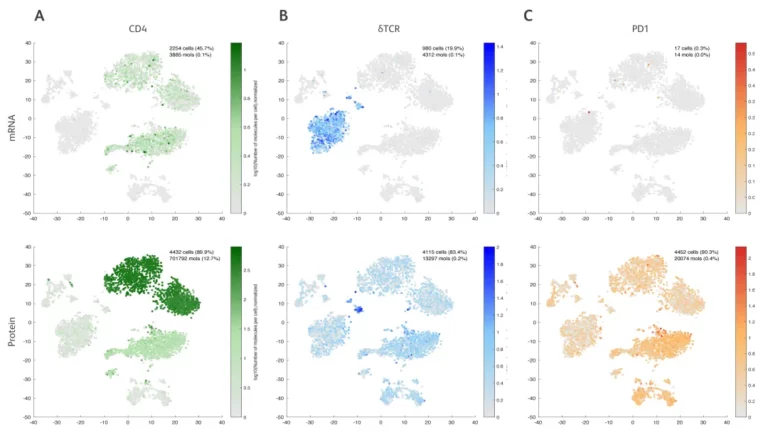
Abseq can reflect protein expression more precisely and overcome the limitations of mRNA detection. For example:
The CD4 – mRNA data shows that it is expressed in both monocytes and CD4+ T cells (upper part of Figure A). The BD AbSeq data shows that it is highly expressed in CD4+ T cells (lower part of Figure A).
The γδTCR – mRNA data shows that it is expressed in both NK cells and γδ T cells (upper part of Figure B), while the BD AbSeq data shows that only a small number of γδ T cells are highly expressed (lower part of Figure B).
The PDL1 – mRNA data shows that it cannot be detected (upper part of Figure C), while the BD AbSeq data shows that it is more highly expressed in certain cell types and specific single cells (lower part of Figure C).
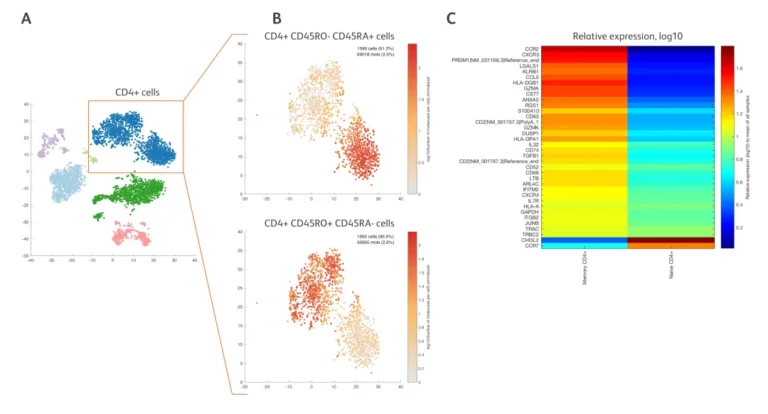
CD45 undergoes splicing at the 5′ end to generate CD45RA and CD45RO, which cannot be distinguished by scRNA-seq. However, the BD AbSeq data shows that there are two different subpopulations (boxed areas) among CD4+ T cells, namely CD4+ naive T cells (Figure 4B, upper panel) and CD4+ memory T cells (Figure 4B, lower panel). Moreover, 36 differentially expressed mRNAs (such as CCR2, CHI3L2) have been identified and are significantly enriched in one of the two cell populations (Figure C).
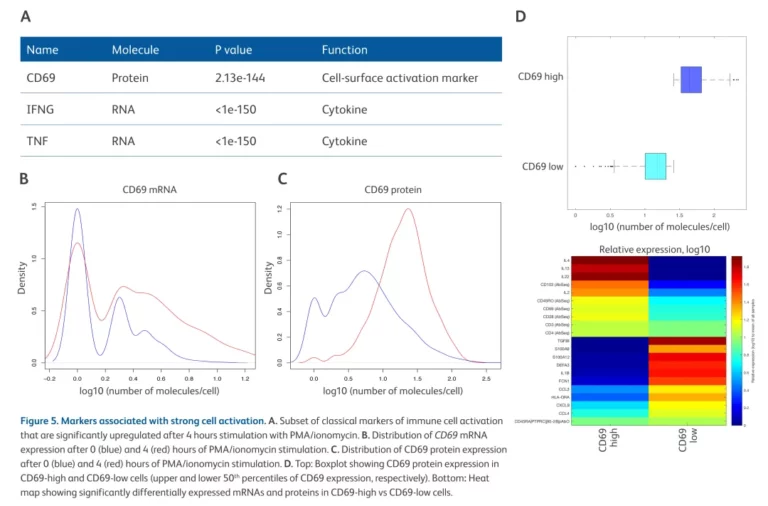
AbSeq was used to study the early responses of immune cells. It was found that after 4 hours of stimulation, IFNγ, TNF mRNA and CD69 protein were significantly upregulated (Figure A), and there were differences in the regulation of CD69 protein and mRNA (Figure B and Figure C). By comparing cells with high and low expression of CD69, more than 20 protein and mRNA markers were identified (Figure D).
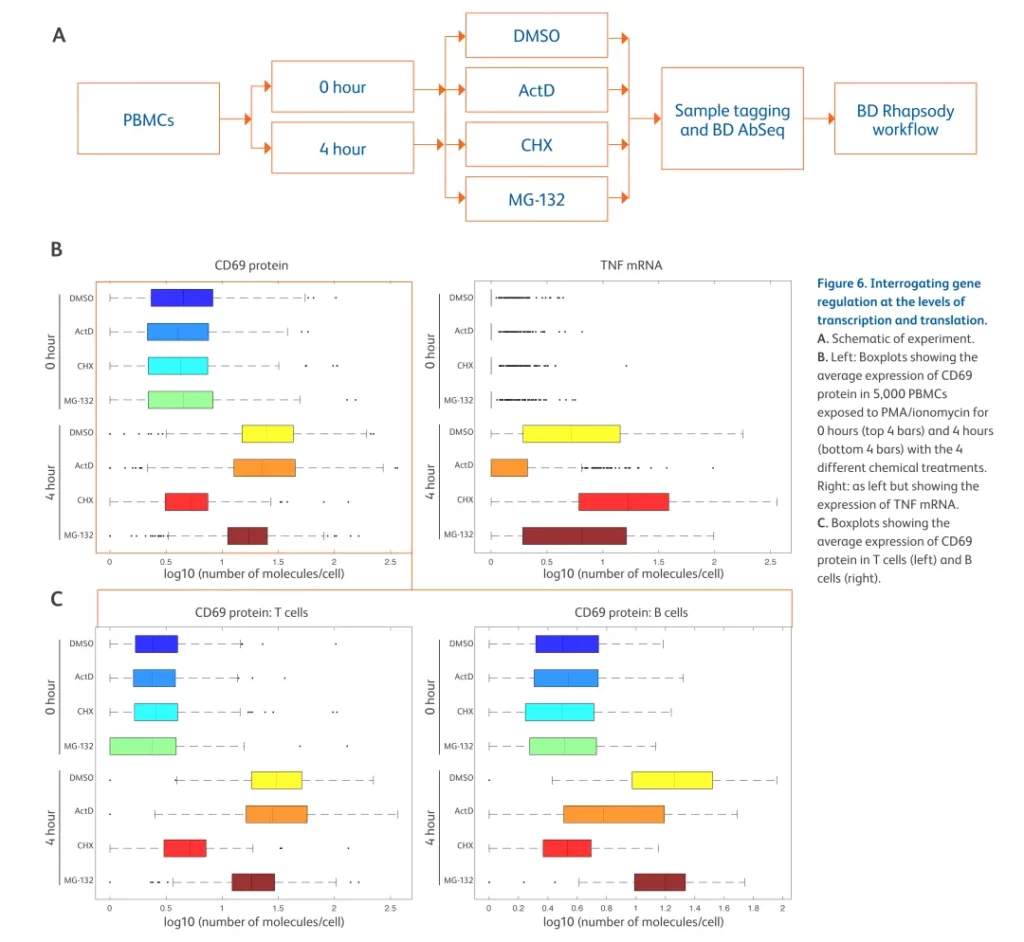
Different inhibitors were used to block transcription, translation, and proteasome activity in stimulation-activated peripheral blood mononuclear cells (PBMCs) (Figure A). The results showed that the expression of TNF mRNA was regulated at the transcription level, while the expression of CD69 protein was regulated at the translation level (Figure B). In addition, the expression of CD69 protein was regulated at the translation level in T cells and was regulated by both transcription and translation in B cells (Figure C).