RXBio Translates Sequence to Science and Industry
Tel: 027-87050299Email: sales@rxbio.cc
- Home
- Single-cell Sequencing
- Spatial Transcriptomics
- Third-generation Sequencing
- Omics Technologies
- Bioinformatics
- Experimental Platform
- About US
中文
RXBio Translates Sequence to Science and Industry
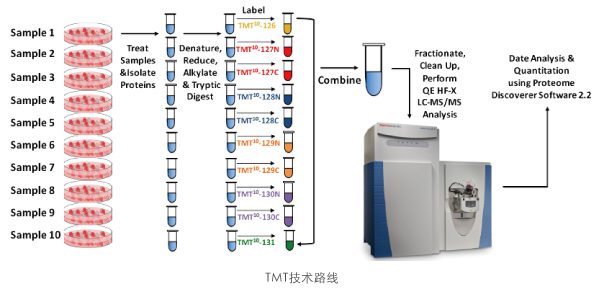
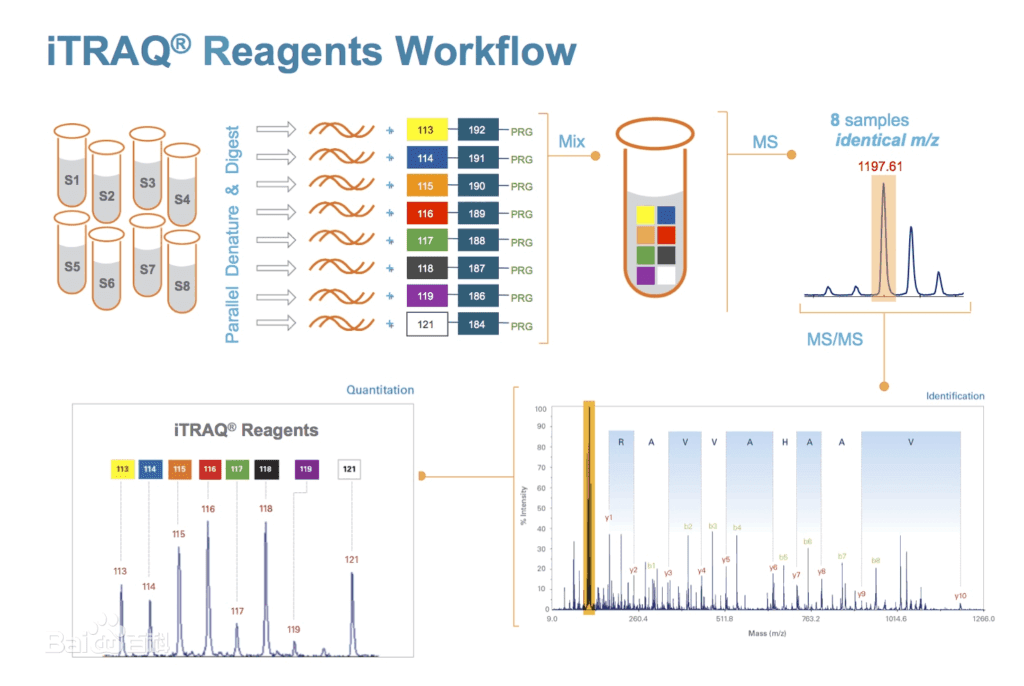
iTRAQ (Isobaric Tags for Relative and Absolute Quantitation) and TMT (Tandem Mass Tags) are in vitro peptide labeling and quantification techniques developed by AB Sciex and Thermo in the United States respectively. Their principle is to use 8 or 10 kinds of isotope tags to label the peptide segments in different samples. After equal mixing, mass spectrometry detection is carried out. By specifically labeling the amino acid groups of polypeptides, relative quantification of proteins and peptide segments in 8 or 10 different samples can be achieved in one mass spectrometry run. They are commonly used high-throughput screening techniques in quantitative proteomics in recent years. They are typically applicable to the quantitative analysis of the whole proteome of cells before and after stimulation, pathological and healthy tissues, and cancer and adjacent tissues.
✔ High accuracy.
✔ No missing values in quantification and good reproducibility.
✔ High throughput.
✔ Applicable to protein quantification of complex samples, such as tissues and cells.
✔ Up to 11 samples can be quantified at one time.
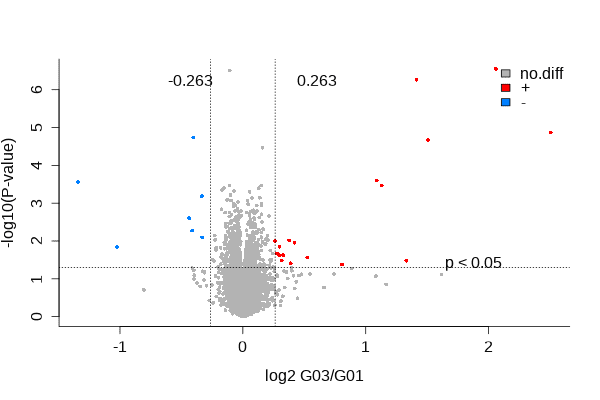
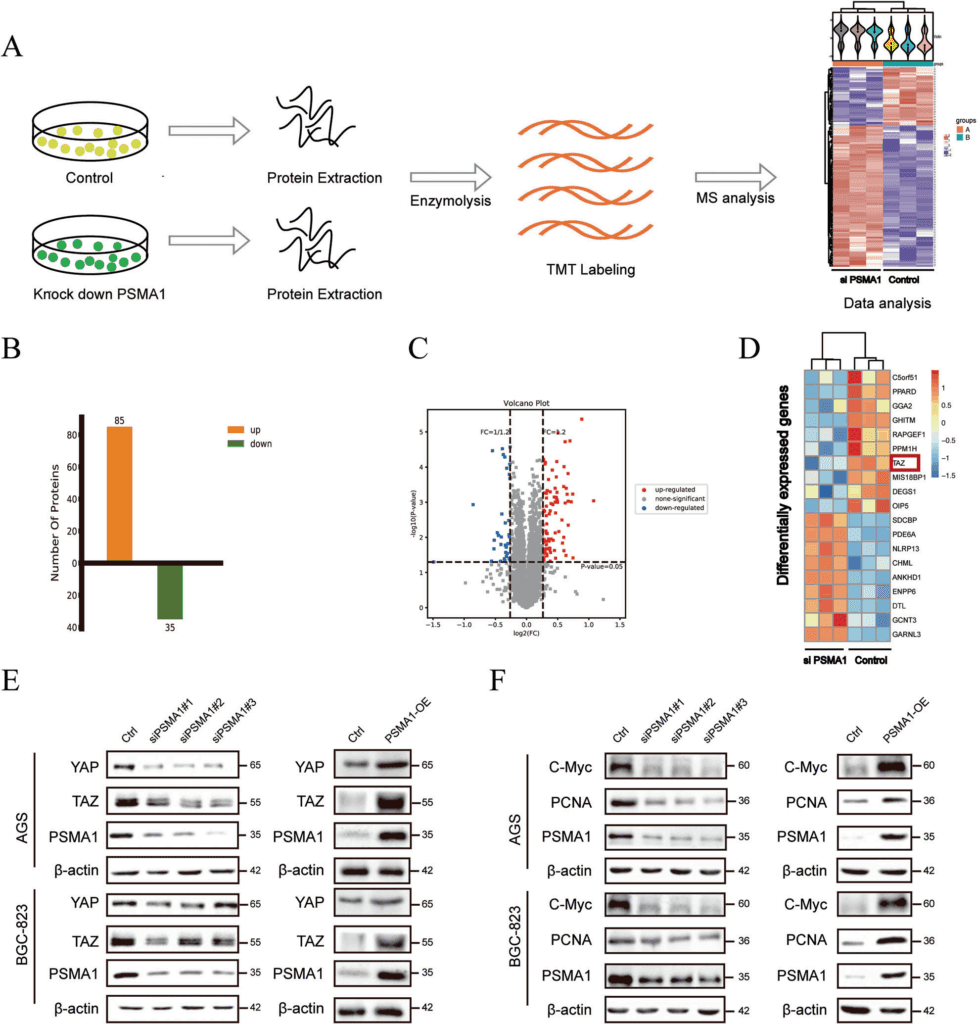
To detect specific substrate proteins that might act as potential interaction targets of PSMA1, the researchers conducted Tandem Mass Tags (TMT) labeled proteomics experiments to systematically detect the changes in the protein profile expression in PSMA1-knockout B-Gastric Cancer-823 cells compared with the control group. The proteomics results showed that PSMA1 siRNA significantly decreased the protein levels of YAP and TAZ.