introduction
DNA pull-down is a technique for studying the interaction between target DNA fragments and proteins under in vitro conditions. DNA pull-down focuses on the DNA sequence of interest and looks for proteins that bind to this sequence. The most common application is to verify the interaction between promoters and transcription factors.
Firstly, biotin-labeled DNA probes are prepared in vitro. The DNA probes can bind to streptavidin magnetic beads. Then, the DNA probe-magnetic bead complexes are incubated with the cell lysate, so that the DNA-binding proteins are adsorbed onto the magnetic beads together. After washing away the unbound proteins, the DNA-binding proteins are eluted from the magnetic beads with an elution buffer to obtain the DNA pull-down products. The DNA-protein complexes can be detected for known proteins by the Western Blot method or identified for unknown proteins by mass spectrometry.
advantages
✔ The probe length can be designed within a wide range.
✔ Proteins are extracted from fresh tissues and cells, maintaining their natural conformations.
✔ It has high specificity and a low false positive rate.
✔ It is applicable to both eukaryotes and prokaryotes.
✔ The experimental cycle is short, with high throughput and high sensitivity.
✔ It enables the enrichment analysis of low-abundance proteins.
✔ The obtained specific proteins are compatible with downstream protein detection technologies.
applications
1. Research on Gene Regulation
The DNA pull-down technique can be used to study the binding between transcription factors and gene promoters, revealing gene regulation mechanisms and transcriptional regulatory networks. This is of great significance for understanding gene expression regulation, developmental processes, and disease progression.
2. Research on Signal Transduction Pathways
The DNA pull-down technique can be used to study the interaction between key proteins and DNA in signal transduction pathways, such as activators, inhibitors, etc., which helps to clarify signal transduction mechanisms and regulatory networks.
3. Research on Disease Mechanisms
The DNA pull-down technique can be used to study the binding proteins of disease-related gene mutation sites or regulatory elements, helping to reveal the occurrence and development mechanisms of diseases and providing a theoretical basis for disease diagnosis and treatment.
4. Drug Development and Screening
The DNA pull-down technique can be used to screen small molecule compounds or drugs that bind to specific DNA sequences, helping to find new drug targets and develop potential drug therapies.
5. Comparative Genomics Research
The DNA pull-down technique can be used to compare DNA-protein interactions among different species, different tissues, or under different conditions, helping to understand changes in genome evolution, tissue specificity, and environmental adaptation.
workflow
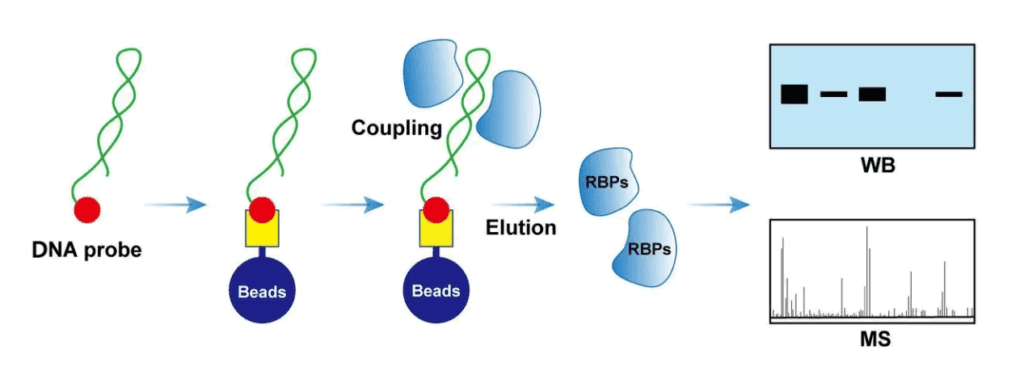
1) Preparation of DNA probes;
2) DNA pull-down experiment: Streptavidin magnetic beads are used to capture DNA probes to form DNA probe-magnetic bead complexes, and then these complexes are incubated with cell lysate for protein capture;
3) Detection and analysis of proteins: Verification by Western Blot and mass spectrometry.
Research Cases
Case 1
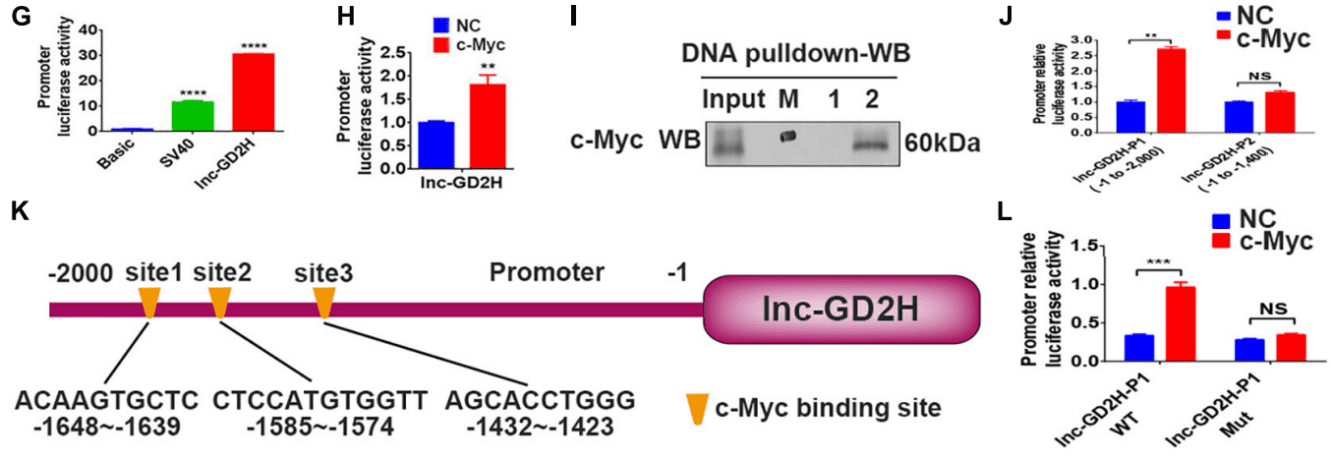
In this study, through experiments such as dual-luciferase assay, chromatin immunoprecipitation (ChIP), DNA pull-down, RNA pull-down, and RNA immunoprecipitation (RIP), the role of lnc-GD2H in muscle regeneration was discovered. During the proliferation of myoblasts, the c-Myc protein binds to the promoter of lnc-GD2H and regulates its transcription, thus promoting the proliferation of myoblasts. During the differentiation of myoblasts, lnc-GD2H interacts with the myoblast differentiation-related protein NACA, inhibits its enrichment at the Myog promoter, alleviates the inhibitory effect of NACA on Myog, and promotes the expression of Myog and the differentiation of myoblasts.
Case 2
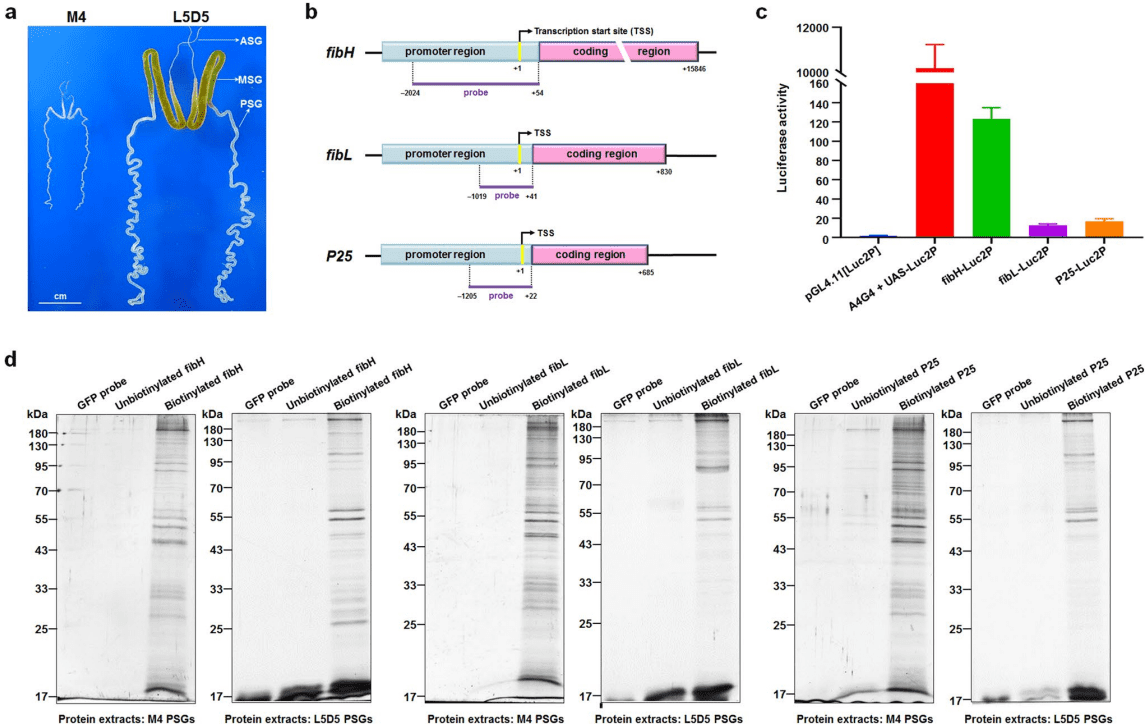 This study reported the promoter-interacting proteins (PIPs) that are essential for regulating the fibroin-encoding genes in the silkworm, including fibroin heavy chain (Fibre H), fibroin light chain (FiBL), and a 25-kD polypeptide protein (P25). The authors identified the proteins that interact with the promoter regions of FibH, FiBL, and P25 through DNA pull-down combined with mass spectrometry analysis. For the first time, this study provided an in-depth map of the proteins that interact with the fibroin gene promoters, which helps to better understand the regulatory mechanisms of silk protein synthesis.
This study reported the promoter-interacting proteins (PIPs) that are essential for regulating the fibroin-encoding genes in the silkworm, including fibroin heavy chain (Fibre H), fibroin light chain (FiBL), and a 25-kD polypeptide protein (P25). The authors identified the proteins that interact with the promoter regions of FibH, FiBL, and P25 through DNA pull-down combined with mass spectrometry analysis. For the first time, this study provided an in-depth map of the proteins that interact with the fibroin gene promoters, which helps to better understand the regulatory mechanisms of silk protein synthesis.
Reference
[1]Chen R. et al: Lnc-GD2H Promotes Proliferation by Forming a Feedback Loop With c-Myc and Enhances Differentiation Through Interacting With NACA to Upregulate Myog in C2C12 Myoblasts. Front Cell Dev Biol. 2021, IF=6.684.
[2]Ma Y. et al: New insights into the proteins interacting with the promoters of silkworm fibroin genes. Scientific Reports. 2021, IF=4.379.
