introduction
SMART-seq (Switching mechanism at 5’ end of the RNA transcript) is a single-cell transcriptome sequencing technology developed in 2012. Its principle is to introduce a customized primer during the reverse transcription process to synthesize the full-length of RNA transcripts within cells, enabling the acquisition of more detailed and comprehensive gene expression data at the single-cell level.
In 2013, SMART-seq2 was reported in the journal Nature Methods. This technology is a sequencing method improved from SMART-seq to enhance the efficiency and coverage of single-cell RNA sequencing. The primer design of SMART-seq2 is more precise, which can better handle low-quality RNA samples and is more sensitive when capturing gene expression information.
advantages
✔ Full-length transcript sequencing: Obtain the complete transcript information of cellular genes and accurately understand the gene expression profiles of single cells.
✔ Wide applicability: Applicable to various cell types and samples, including low-quality single-cell RNA.
✔ Single-cell resolution: Possess high single-cell resolution and be able to deeply explore cell heterogeneity and subtypes.
✔ Low-input RNA samples: Require a lower amount of RNA input and are suitable for samples with limited cells.
workflow
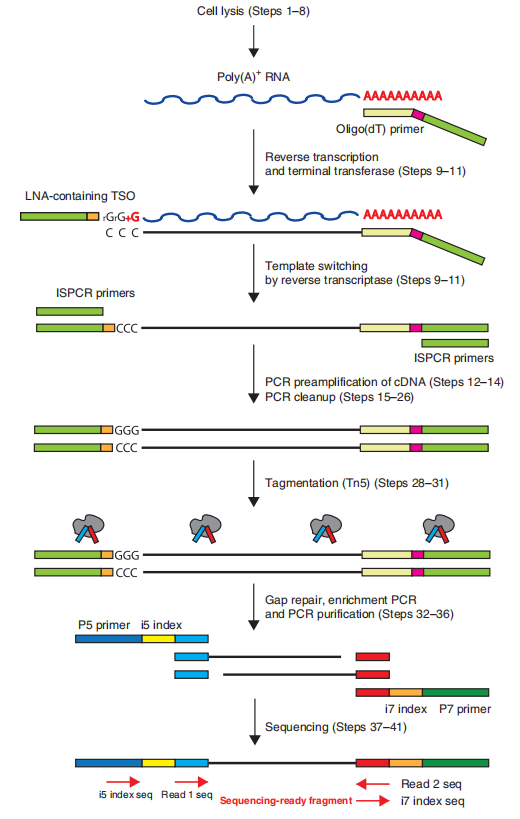
This technology uses oligo(dT)VN Primer as the reverse transcription primer. By taking advantage of the template-switching activity of MMLVRT, a linker sequence is added to the 3’ end of the cDNA. Reverse transcription is carried out through this linker sequence to generate the first strand of cDNA.
When the reverse transcriptase reaches the 5’ end of the mRNA, it will continuously add several cytosine (C) residues at the end. Then the TSO primer is added. After annealing, it binds to the 3’ end of the first strand and hybridizes with the poly(C) overhang to synthesize the second strand.
The resulting cDNA is amplified by PCR to obtain nanogram-level DNA, which can be used for sequencing after purification.
Research Cases
Case 1
In 2020, researchers from Huashan Hospital of Fudan University in Shanghai, East Hospital Affiliated to Tongji University, Auburn University in the United States and other institutions published an article titled “Circulating tumor cell characterization of lung cancer brain metastases in the cerebrospinal fluid through single-cell transcriptome analysis” in the journal “Clinical and Translational Medicine”.
The researchers established an effective method for collecting cerebrospinal fluid circulating tumor cells (CSF-CTCs) and isolated single cerebrospinal fluid cells from 5 patients with leptomeningeal metastases of lung adenocarcinoma (LUAD-LM) and 3 control groups. They used the Smart-seq2 technology to conduct single-cell sequencing on 3,792 cells and comprehensively characterized the gene expression of cerebrospinal fluid cells. After filtering out low-quality cells, the article analyzed 207 normal cerebrospinal fluid cells, 41 B cells and 41 T cells. On average, 803 gene expressions were detected in each cell. Cluster analysis revealed that normal lymphocytes had similar expression profiles under different microenvironments. No B cells were found in normal cerebrospinal fluid samples.
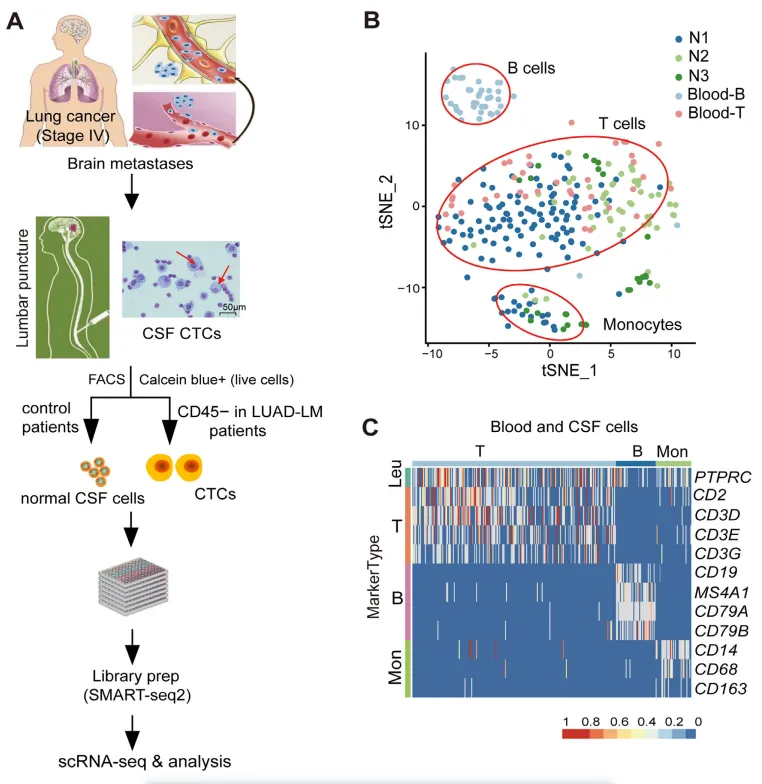
Figure 1. Isolation of cerebrospinal fluid circulating tumor cells (CFS-CTCs) and the composition of normal cerebrospinal fluid cells.
Case 2
On March 2, 2021, the team led by Professor Zhang Zemin from Peking University published a research paper titled “Direct Comparative Analyses of 10x Genomics Chromium and Smart-seq2” in Genomics Proteomics Bioinformatics. In this study, two single-cell transcriptome sequencing technologies, Smart-seq2 and 10x Genomics Chromium, were used to conduct a comparative analysis of CD45- cells sorted by flow cytometry in liver cancer (LT) and its adjacent non-tumor (NT) tissues from a patient with hepatocellular carcinoma (HCC), as well as in rectal cancer (PT) and liver metastasis (MT) tissues from a patient with rectal cancer accompanied by liver metastasis. Differences between the two were analyzed from multiple perspectives, providing a basis for the selection and application of single-cell transcriptome sequencing technologies.
The dropout rate in 10x was higher than that in Smart-seq2. Taking the HK gene in LT cells as an example, the dropout rate in 10x was also higher than that in Smart-seq2. Genes with lower expression levels had higher dropout rates. In 10x, genes with lower abundances were detected from a smaller number of cells, and these genes might lead to higher noise. The authors also found that the coefficient of variation (CV) of gene expression was related to the dropout rate (CV is often used to measure the expression variation of a gene in certain samples). Genes with larger CVs usually had lower expressions, especially for 10x. Meanwhile, genes with larger CVs also had higher dropout rates.
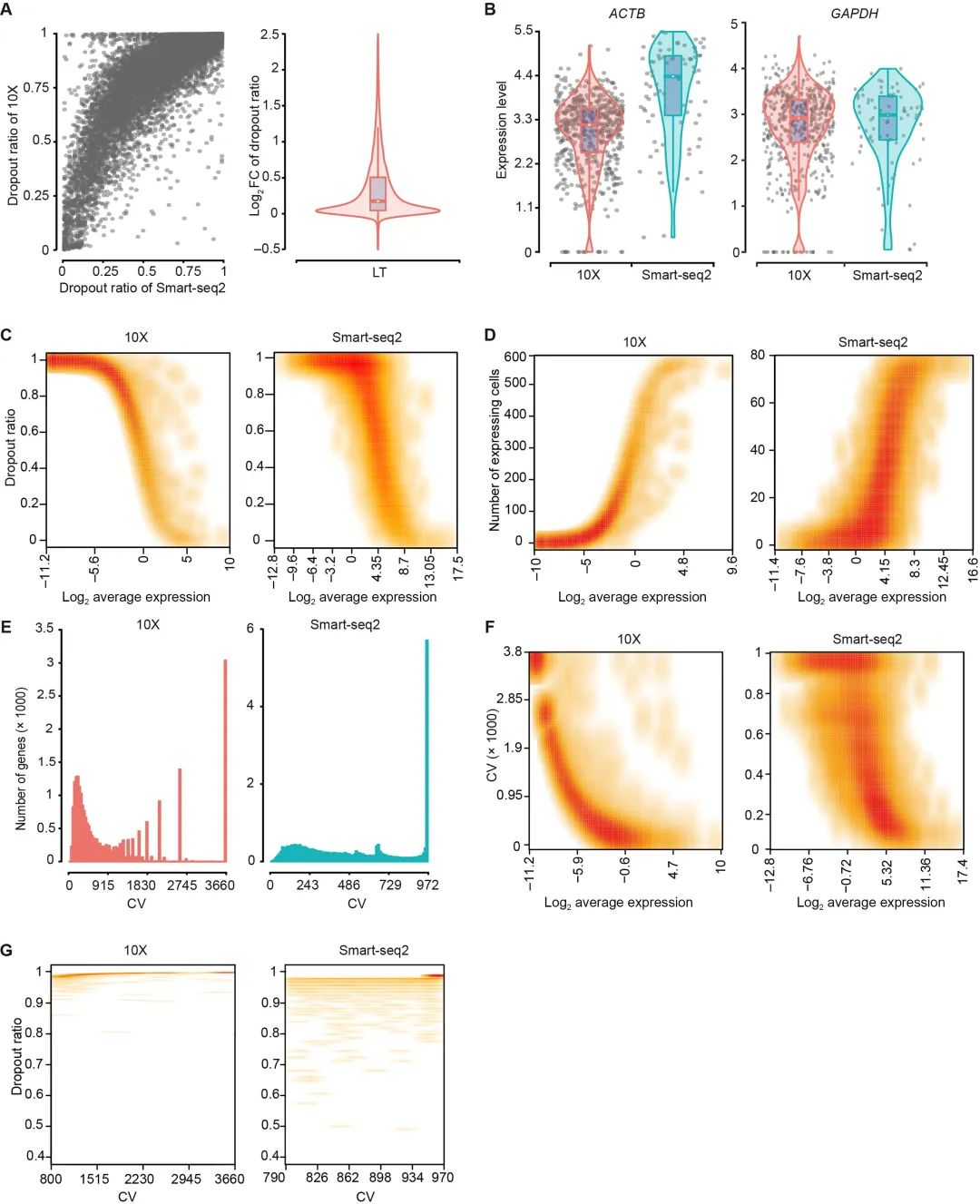
Figure 2 Results of Dropout Evaluation for LT Cells
