Introduction
Second – generation transcriptome sequencing cannot provide the results of quantitative analysis for multiple transcripts of a gene respectively, because this technology is not suitable for such a situation. A single read of second – generation short sequencing cannot span the full – length transcript. It is necessary to use software to splice short reads to obtain transcripts, and then evaluate the expression level. There may be problems of splicing errors or incompleteness, so the complete transcript cannot be accurately obtained, and the source of reads of multiple transcripts of a gene cannot be distinguished. There is a phenomenon of multiple alignments of short reads in highly conserved regions with high similarity among different genes, which leads to inaccurate or biased quantitative expression at the transcript or even gene level. In contrast, Nanopore third – generation full – length sequencing does not require breaking and splicing transcripts. One read can span the full – length transcript, and the multiple alignment rate is low, so it can accurately obtain the respective expression information of multiple full – length transcripts of a gene. From second – generation transcriptome to Nanopore third – generation full – length transcriptome, platform upgrading and technological innovation have been realized, which can solve the problems that cannot be solved by second – generation sequencing.
However, compared with second – generation sequencing technology, the length of each read in third – generation sequencing technology is significantly longer. When the amount of sequencing data is the same, the number of reads generated by third – generation sequencing will be significantly less than that by second – generation sequencing. For some transcripts with low expression levels, this requires extremely high sequencing depth to achieve effective detection. For example, when performing saturation detection of the human transcriptome, more than 30 Gb of sequencing data is required. Such high requirements will inevitably greatly increase the cost of third – generation sequencing and bring a greater economic burden to research work. Targeted sequencing technology enables researchers to screen out gene loci of interest, thereby reducing sequencing costs and workload, so as to obtain data with high coverage in target regions.
Oxford Nanopore Technologies Targeted Long – Read Sequencing (ONT – TlrRNA – seq) is to amplify the full – length region of the transcript of interest by PCR, and then directly sequence the amplicons.
The core advantage of ONT – TlrRNA – seq technology is that it can perform deep sequencing for specific transcripts without comprehensively sequencing the entire transcriptome. This method not only significantly improves the sequencing efficiency, but also enables researchers to focus more on transcripts directly related to diseases or specific biological processes.
Through ONT – TlrRNA – seq technology, researchers can quickly identify the expression of transcripts of key genes, providing strong support for subsequent transcript function research and disease diagnosis. In addition, ONT – TlrRNA – seq technology is also widely favored because of its high flexibility and simple operation, and has become an important technology in the current field of transcript detection.
Advantages
✔ The sequencing length ranges from thousands to tens of thousands of base pairs, covering the full length.
✔ There is no need for splicing, and it is naturally full-length.
✔ It can directly identify transcript isoforms.
✔ It enables precise quantification and can also detect transcripts with low expression.
✔ The amount of sequencing is low, and it offers high cost-performance.
Research Cases
Case 1
Title: Patients with Asian-type DEL can safely be transfused with RhD-positive blood
Journal: Blood
Impact factor: 21
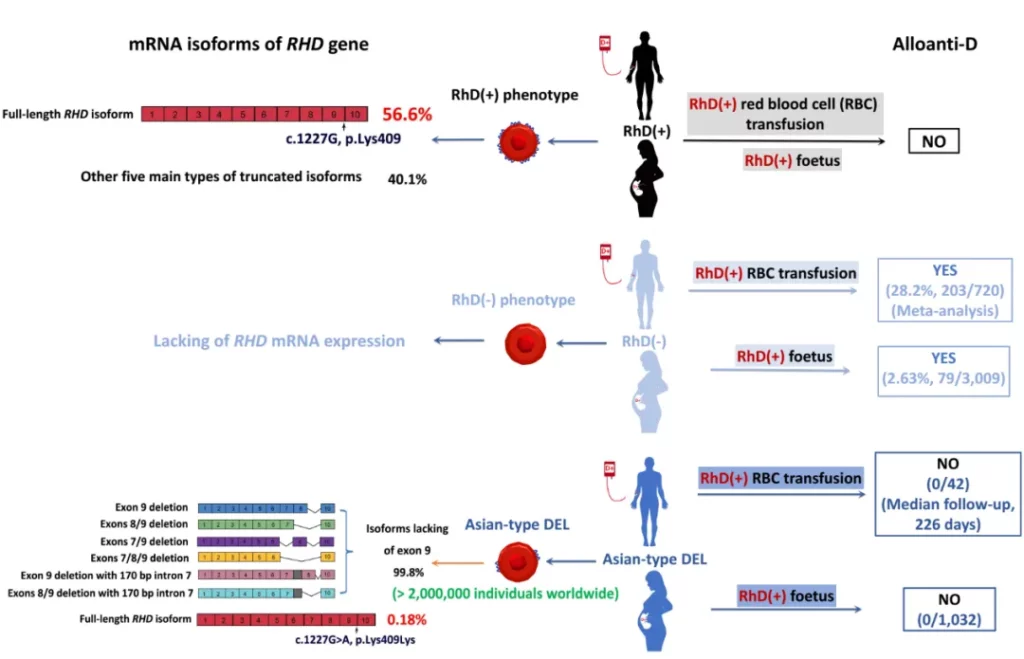
This study has provided strong evidence for the safe transfusion of RhD-positive blood to patients with the “Asian-type” DEL blood group from three aspects.
Firstly, a multicenter clinical trial on the transfusion of RhD-positive red blood cells to patients with the “Asian-type” DEL blood group was carried out for the first time. Through long-term follow-up, it was found that patients with the “Asian-type” DEL blood group could safely receive RhD-positive red blood cells without the occurrence of anti-D alloimmunization.
Secondly, a retrospective study on pregnant women with the “Asian-type” DEL blood group was conducted nationwide. Large-sample data confirmed that pregnant women with the “Asian-type” DEL blood group would not develop anti-D alloimmunization when carrying an RhD-positive fetus, which was statistically different from pregnant women with truly RhD-negative “panda blood”. Meanwhile, among pregnant women who had already developed alloanti-D, no pregnant women with the “Asian-type” DEL blood group were found.
Finally, with the help of the third-generation Nanopore sequencing technology, the full-length transcript of the RHD gene carrying the 1227A mutation was detected for the first time in the nucleated red blood cells of individuals with the “Asian-type” DEL blood group. In vitro experiments confirmed that it could express the RhD antigen with a complete epitope, which was consistent with the antigenic epitope of RhD-positive red blood cells. This indicates that patients with this blood group may avoid the occurrence of alloimmunization through immune tolerance.
The above research results have provided extremely powerful evidence for formulating clinical transfusion guidelines for the routine transfusion of ordinary RhD-positive blood to patients with the “Asian-type” DEL blood group, and can further solve the problem of blood supply difficulties for patients with this blood group.
In addition, pregnant women with the “Asian-type” DEL blood group do not need to be injected with anti-D immunoglobulin, nor do they have to frequently monitor anti-D alloimmunization during pregnancy. This can not only save the precious blood product of anti-D immunoglobulin but also save tens of millions of yuan in injection and monitoring costs of anti-D immunoglobulin for pregnant women with this blood group every year. The research results can also benefit the “Asian-type” DEL blood group population in Japan, South Korea, Southeast Asian countries, and developed countries with a large number of East Asian immigrants.
Case 2
Title: Discovery of circular transcripts of the human BCL2-like 12 (BCL2L12) apoptosis-related gene, using targeted nanopore sequencing, provides new insights into circular RNA biology
Journal: Funct Integr Genomics
Impact factor: 3.9
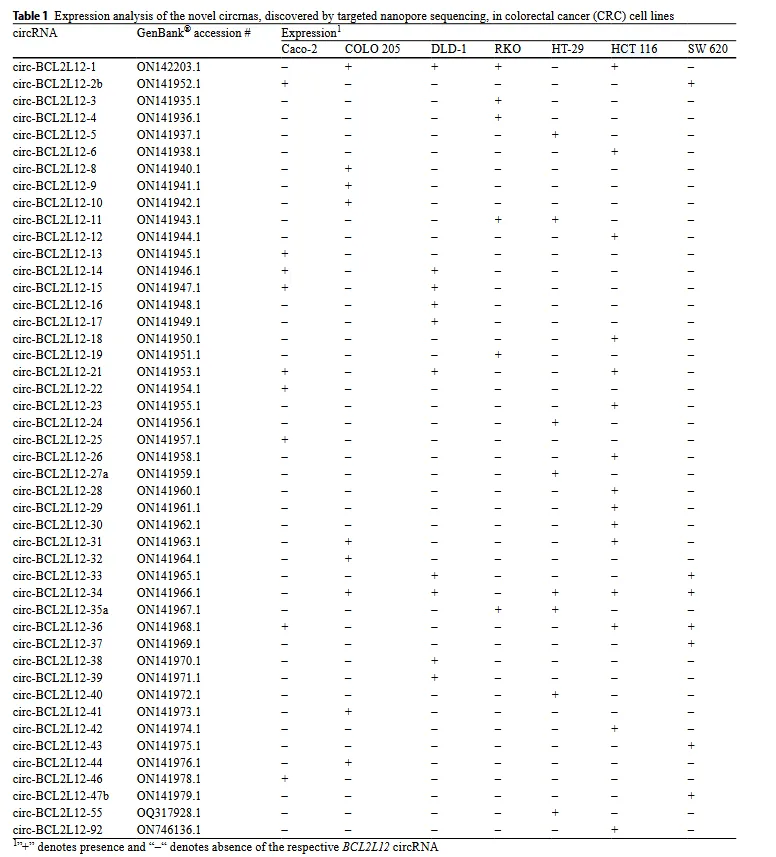
The researchers designed two pairs of divergent primers targeting each annotated exon of BCL2L12 and carried out nested PCR. After mixing and purifying the PCR products of different cell lines, a nanopore sequencing library was constructed. Then, bioinformatics analysis was conducted using publicly available tools and self-developed algorithms, and a new process for identifying novel circRNAs was successfully developed. In CRC (colorectal cancer) cell lines, the researchers identified a total of 46 BCL2L12 circRNAs, most of which were discovered for the first time. These circRNAs covered approximately half of the region of the BCL2L12 gene, and there was only a small overlap compared with the circRNAs recorded in existing public databases.
In addition, some of the circRNAs were composed of a single exon, and others contained exons that had never been described before. The frequencies and coverages of these exons exhibited unique characteristics. Through the analysis of nanopore sequencing data, the researchers found that there were significant differences in the expression of BCL2L12 circRNAs among seven CRC cell lines. The exon structures and intron coverage of circRNAs in different cell lines were not the same. Among them, exons 4 and 5 had a relatively high frequency of occurrence in circRNAs, but their specific structures in circRNAs of different cell lines were different.
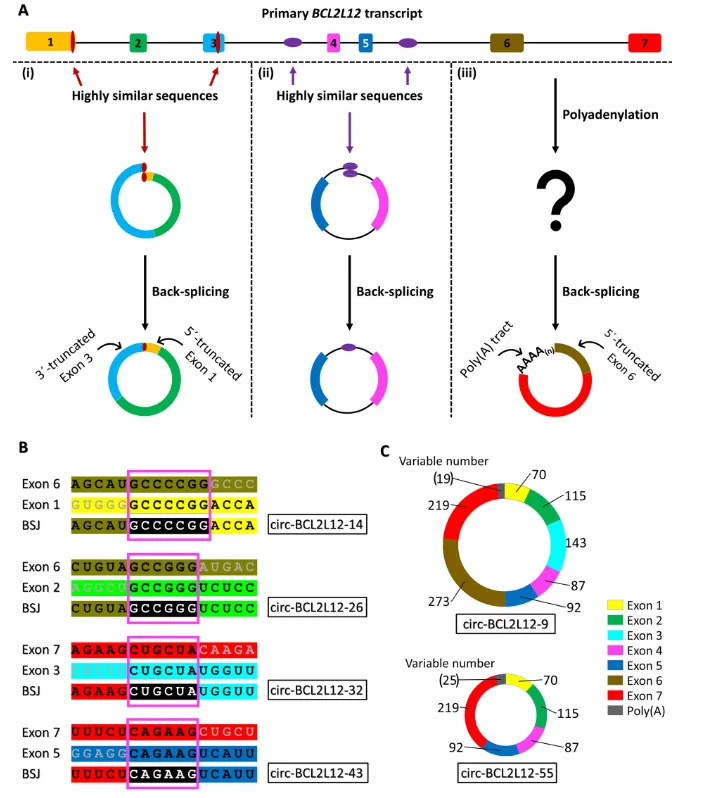
Among the 46 identified circRNAs, the researchers observed 40 different back-splicing junctions (BSJs). The back-splicing junctions of most circRNAs did not conform to the norm, and in most cases, the precursor mRNA regions that formed the BSJs had a high degree of sequence similarity. In addition, the study also found that two circRNAs (circ-BCL2L12-9 and circ-BCL2L12-55) contained poly(A) fragments similar to the poly(A) tail of messenger RNA (mRNA), and this finding challenges the traditional understanding of circRNAs.
Reference
1.Ji, Y.,Luo, Y.,Wen, J.,Sun, Y.,Jia, S.,Ou, C.,Yang, W.,Chen, J.,Ye, H.,Liu, X.,Liang, Y.,Lu, Z.,Feng, Y.,Wu, X.,Xiao, M.,Mo, J.,Zhou, Z.,Wang, Z.,Liao, Z.,Chen, J.,Wei, L.,Luo, G.,Santoso, S.,Fichou, Y.,Flegel, W.A,Shao, C.,Li, C.,Zhang, R., & Fu, Y. (2023). Patients with Asian-type DEL can safely be transfused with RhD-positive blood. Blood, 141 (17), 2141-2150.
https://doi.org/10.1182/blood.2022018152
2.Karousi, P., Kontos, C.K., Nikou, S.T. et al. Discovery of circular transcripts of the human BCL2-like 12 (BCL2L12) apoptosis-related gene, using targeted nanopore sequencing, provides new insights into circular RNA biology. Funct Integr Genomics 25, 66 (2025). https://doi.org/10.1007/s10142-025-01578-1
